Context
Organic solid waste is emerging as an attractive renewable resource for the production of biofuels and synthons through anaerobic bioprocesses. However, waste matrix is complex, heterogeneous, and temporally variable and such features contribute to the complexity of their valorisation: organic waste biorefinery is currently in the exploratory research phase. To orientate fermentation pathways and establish stable processes, new operational levers are required. Since host-virus relationships are highly specific, viruses could serve as the basis for the development of biocontrol tools specifically targeting certain functional microbial groups. In other sectors, including the medical field (phage therapy), agro-food-sector and waste water treatment, viruses and their components are indeed already used or considered for biocontrol applications. Regarding waste biorefinery, methanogens are particularly detrimental to the production of molecules more valuable than methane (e.g. ethanol, butanol, lactate…) and are a relevant target for biocontrol. However, viruses of methanogenic archaea are still poorly characterized so that the development of such strategies is currently not possible.
Objectives
The project VIRAME thus aims at characterizing in situ the genomic content of viruses infecting methanogens within anaerobic bioprocesses for organic waste valorisation. Establishing the link between a virus and its host within ecosystems is challenging since they do not systematically share common genes. To overcome this bottleneck, the link between activated methanogenesis pathways, archaea catalysing methanogenesis and the genomic content of viruses infecting these archaea will be established thanks to an original integrated approach coupling state-of-the-art molecular ecology tools (in particular meta-omics) and in silico analyses (comparative genomics, bioinformatic approaches specific to viral metagenomics).
Work packages
The project VIRAME is organised into 4 work packages shown on the figure below.
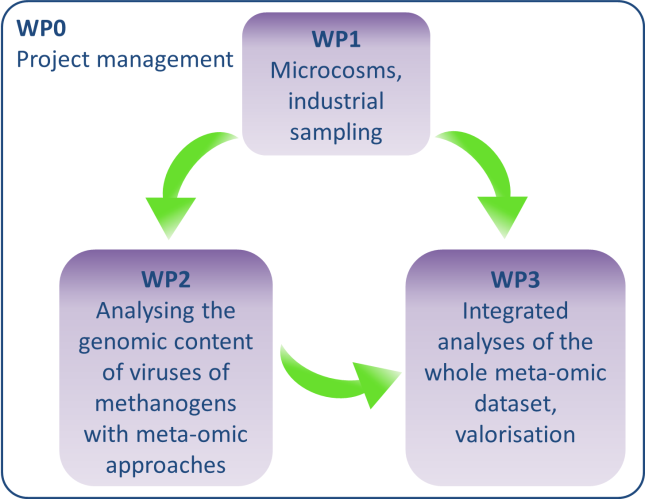
Viruses of organic waste anaerobic digestion ecosystems will be obtained from laboratory microcosms designed to activate various methanogenic populations as well as from industrial samples (WP1). The nucleic acids from virions and cell populations will be extracted, sequenced and analysed, to identify and study sequences originating from viruses of methanogenic archaea (WP2). Integrated analyses (WP3) of the whole dataset will enable to considerably extend the knowledge on the diversity of viruses from methanogenic archaea. Strategies for the biocontrol of methanogens will moreover be suggested.
